All published articles of this journal are available on ScienceDirect.
New Insights into the Interaction of Free-Living Amoebae and Pandoravirus Inopinatum: Investigations of the Host Range and the Role of Multilamellar Bodies
Abstract
Objective:
FLA are predatory heterotrophic microorganisms, feeding as trophozoites on bacteria, cyanobacteria, fungi and algae while adhering to surfaces through phagocytosis. It is known that FLA produce and expel vesicles as part of the digestive process. Bacteria are packaged in multilamellar bodies and are released into the environment if not digested. In 2008, it was shown how easily the so-called Pandoraviruses can get in close contact with humans, while proliferating in Acanthamoeba.
Materials and Methods:
In our study, the search for these packages and multilamellar bodies in Acanthamoebae was conducted by electron microscopy with special emphasis on Pandoravirus inopinatum virions being involved in these processes. The multilamellar bodies were detected within the trophozoites of the amoeba host strain and as expelled vesicles within their environment. Neither digested, partially digested or viable Pandoravirus inopinatum virions nor developmental stages were found within these packages. A modified host range evaluation method was used to study the uptake and the proliferation of Pandoravirus inopinatum virions by Acanthamoeba trophozoites via light microscopy and to determine the host range.
Results:
In addition to the Acanthamoeba strain, which was found to harbor Pandoravirus inopinatum initially, we confirmed another 9 Acanthamoeba strains to be susceptible, among them members of genotypes T4 and T 11.
Conclusion:
The modified time series method, which we used, proved to be superior to the initial (more static) host range studies, in both axenic and xenic cultures.
1. INTRODUCTION
Naked free-living amoebae (FLA) graze biofilms and feed on bacteria, algae, yeasts and other protozoa via phagocytosis following chemotactic trophism [1-3]. They capture their prey by phagocytosis and transfer them to lysosomal compartments in the phagocytic pathway where they are usually digested by enzymes [4, 5].
However, some of these intracellular microorganisms (endocytobionts) have developed a strategy to avoid lysis and digestion during the phagocytic process. These endocytobionts escape digestion by host cell lysis or exocytosis as free microorganisms ( e.g. bacteria) or packaged in the excreted/egested pellets [5, 6]. The impact of such interactions of FLA and their endocytobionts with respect to Public Health and Environmental Health was described recently [7]. The relevance of the FLA - endocytobiont relationship in terms of pathogenicity, tenacity, virulence enhancement, protection, gene transfer etc. is within the focus of current research [8].
In 2008 an extraordinary endocytobiont was detected in Acanthamoebae, isolated from the contact lens storage case of an Acanthamoeba keratitis (AK) patient [9]. Due to the lack of identification of the phylogenetic affiliation, Pandoravirus inopinatum was first published as being an endocytobiont of the Acanthamoebae [9], the intracellular development and morphological details were addressed subsequently [10]. It was demonstrated that the “extraordinary endocytobionts” were phagocytosed via food cup building after adhesion to the surface of Acanthamoeba trophozoites [10]. The endocytobionts proliferated within the Acanthamoeba trophozoites and consequently destroyed the amoebal trophozoites. In 2015, the endocytobiont was phylogenetically grouped with the so-called Giant viruses (or “giruses”) and named Pandoravirus inopinatum following the detection of two environmental Pandoravirus isolates and the subsequent full genome sequencing [8, 11, 12].
In general, viruses possess no self-reproduction mechanism and require a host cell to use their protein biosynthesis; this is one of the reasons, that they are usually not attributed as belonging to the tree of life. The Pandoraviruses belong to the DNA-viruses with a double strand genome of 1,9-2,5 MBP, which makes them the biggest viruses ever detected with respect to their genome [11]. The genetic investigation of Pandoravirus inopinatum revealed a genome of 2.243.109 base pairs of which 89 and 85% were congruent with the environmental isolates Pandoravirus dulcis and Pandoravirus salinus respectively [12]. This genome size and the enormous size of the virions with >1µm are not typical features attributed to viruses (except giant viruses). One advantage of the enormous size of the virions is the fact that light microscopy can be used to detect and further examine these viruses and their interaction kinetics.
FLA trophozoites produce and expel vesicles as part of their digestive process. These expelled vesicles (also called fecal pellets) vary among the FLA according to the environmental and trophic conditions [13] In the case of FLA, extracellular vesicles containing digested or viable bacteria are known. They are expelled by exocytosis, which is a continuously active process for feeding amoebal tophozoites.
The potential role of FLA which package bacteria in this manner and its importance in the persistence and transmission of pathogenic bacteria was discussed recently [13]. As a number of protozoa, among them the FLA, ingest other microorganisms, package them in membrane structures and subsequently release them into the environment, we searched for these packages and multilamellar bodies in electron microscopic photos taken during the “co-cultivation” assay of Acanthamoeba sp. strain LaHel and Pandoravirus inopinatum. Packaged microorganisms are more resistant to environmental conditions, which could be of relevance in terms of tenacity and protection but also of persistence and transmission of Pandoravirus virions. Therefore, it had to be determined whether the packaging processes and/or the multilamellar bodies are involved in the development (or the egestion) of Pandoravirus inopinatum at any time.
While the development of Pandoravirus inopinatum was already examined using electron microscopy, light microscopy, staining etc. a thorough and detailed investigation of the host spectrum was lacking to date [9, 10]. Therefore, an enhanced host spectrum analysis was initiated using different Acanthamoeba strains from the laboratory as potential hosts of Pandoravirus inopinatum. We intended to identify the susceptibility of different Acanthamoeba strains and their development dynamics with respect to the proliferation of Pandoravirus inopinatum. These host spectrum analyses and further details of the uptake and replication were conducted in a modified manner using time series investigation and prolonged examination times in contrast to standard (more static) host range analyses. Xenic and axenic culture methods were compared with respect to different variables. Using the time series investigation, detailed differences between the interactions of the Acanthamoeba strains and the Pandoravirus should be highlighted. The uptake of the virus was examined thoroughly to compare the interaction features quantitatively and chronologically. Putative advantages of this modified host spectrum analysis method had to be separately evaluated.
2. MATERIALS AND METHODS
2.1. Materials and Methods for the Investigation of Multilamellar Bodies
2.1.1. Electron Microscopy
Xenically grown amoebae were extracted after incubation with Pandoravirus inopinatum virions for 3 to 5 days; axenically grown amoebae were extracted after incubation (co-cultivation) for 5 to 7 days. The resulting suspension (Infected amoebae in amoebal saline) was pelleted by centrifugation (10 min: 500G). The resulting pellets were fixed in 3% glutaraldehyde and transferred in 0.1 M cacodylate buffer (pH 7.2) for 1 h, washed twice in the same buffer, postfixed for 1 h in 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.2), and embedded in Spurr resin. Subsequently, the sections were stained with uranyl acetate and Reynold's lead citrate and examined using a Leo EM 910 transmission electron microscope [10]. The resulting photos of several time series were investigated thoroughly for the presence of multilamellar bodies and Pandoravirus particles which were possibly involved in the packaging processes.
2.2. Materials and Methods Used for time Series Investigation
2.2.1. Host Range Study
The host amoebae were taken from the collection of our parasitology lab in Koblenz/Germany. 9 Acanthamoeba strains were used in addition to the “original” host Acanthamoeba sp. strain LaHel. This Acanthamoeba strain LaHel resulted from patient material tested positive for Acanthamoeba keratitis [9]. Optimal growth conditions were provided at 25°C. Growth dynamics, cyst building rate and density of the monolayer (in xenic populations) of the used host amoebae were evaluated in preliminary tests [14, 15]. “Co-cultivation” experiments followed the established methods described earlier [16].
2.3. Xenic Culture of the Host Amoebae
The amoebae were cultured on 1,2% Non-Nutrient-Agar with Enterobacter cloacae (grown in Trypticase-Soya-Bouillon, BD, Heidelberg) as a food source as described in Scheid et al. 2008 and Schroeder 2016. 0,5mL virus (virion) containing suspension was used to provide sufficient material onto a NN-agar plate with the xenically grown test amoebae. The virion suspension was added as soon as the host amoeba population showed a well-grown monolayer (85% confluence) with trophozoites [14]. 6 ml of Amoebic Saline (AS) were added to the xenic cultures with the Acanthamoeba trophozoites harbouring the endocytobionts which were scraped from the agar surface by means of a Drigalski spatula. This suspension was filtered through a 5 µm filter (Sartorius) to separate infectious viral particles from their host amoebae. After centrifugation of the filtered suspension (15 min; 2500 rpm) the pellet was resuspended and co-cultivated with various FLA strains (Table 1), grown on agar plates and incubated at 24°C.
| Host Species | Strain | T-Group | Source | Gen Bank Acc. Nr. |
|---|---|---|---|---|
| Acanthamoeba sp. | LaHel | T4 | contact lenses, keratitis patient | KP236736 |
| Acanthamoeba sp. | TaHa | T4 | contact lenses, contact lens storage case; keratitis patient | KJ476518 |
| Acanthamoeba sp. MaTa | MaTa | T11 | contact lens storage case; keratitis patient | KY569460 |
| Acanthamoeba sp. | MaHa | T4 | contact lens storage case; keratitis patient | KU935612 |
| Acanthamoeba sp. | Petz | T4 | contact lens storage case; keratitis patient | KJ476517 |
| Acanthamoeba sp. | KLB | T4 | contact lens storage case; keratitis patient | KU935611 |
| Acanthamoeba sp. | Ubo | T4 | contact lens storage case; keratitis patient | KY563326 |
| Acanthamoeba sp. | Ulse | T4 | contact lens storage case; keratitis patient | KY498611 |
| Acanthamoeba sp. | Smith | T4 (Grp. II) | contact lens storage case; keratitis patient | KJ476516 |
| Acanthamoeba sp. | FloBö | T4 | contact lens storage case; keratitis patient | KY498610 |
| Acanthamoeba sp. + Pandoravirus inopinatum | LaHel, KLaHel | contact lens storage case; keratitis patient | KP236736, KP136319 |
2.4. Axenic Cultures of the Host Amoebae
The host amoebae were transferred from xenical cultures into axenical cultures in T25 flasks (25cm2 cell culture flasks, Greiner Bio-One GmbH, Frickenhausen, Germany). SCGYE (Chang) medium was used as a result of several pre-studies (after evaluation against Jensen medium) to afford the axenic conditions [17, 18]. 0,5mL virus (virion containing) suspension was used to provide sufficient material in a 24 well plate (Sigma-Aldrich, Munich, Germany) filled with axenically grown trophozoites of the test amoebae [14]. The Pandoravirus inopinatum virions resulted from xenical culture. The suspension was filtered through a 5 µm filter (Sartorius) to separate infectious viral particles from their host amoebae. After centrifugation of the filtered suspension (15 min; 2500 rpm) the pellet was resuspended and co-cultivated with various FLA strains (Table 1), grown axenically in bacteria-free medium and incubated at 24°C.
2.5. Modified Time Series Analysis
Light microscopy and documentation were performed using Leica DM 5000 with a digital camera JVC KY-F1030. In addition, the Zeiss Axioskop 2 plus was used with the digital camera Hitachi KP-D50. The documentation and storage of the microscopic images, time series and videos was performed using the software DISKUS 4.30, Hilgers, Königswinter, Germany. This software enables the user to create (video) records of the interaction during several hours so that detailed insights into the interaction between host and virus can be visualized. To gain results with precise and meaningful details, expedient microscopical photos, especially the optical properties had to be checked and regulated every 30 seconds. To achieve more detailed information about uptake, development and replication rates, the software was used to achieve time series at pre-determined time intervals. This enabled the detailed observation of one trophozoite interacting with a virion (or several virions) during the course of several hours. The microscopic analysis of this one trophozoite was performed in 400x and 1000x magnification for up to 5 hours. These long-term studies led to more than 5000 single photos (>1,5GB) and numerous videos, for each study [14, 15].
3. RESULTS
3.1. Multilamellar Bodies by Acanthamoeba sp. Strain LaHel
During our light microscopic time series studies, a discharge of Pandoravirus virions by trophozoites of the Acanthamoeba sp. strain LaHel, or an exocytosis prior to proliferation was not observed. Therefore, electron microscopic photos of Acanthamoeba sp. strain LaHel and Pandoravirus inopinatum were consulted to search for mutilamellar bodies or exocytosis vesicles with Pandoravirus particles. In the case of Acanthamoeba sp. grown xenically on non-nutrient Agar, with Enterobacter cloacae as a food source (to mimic real environmental conditions in contrast to axenic culture), the packaging was detected and documented (Figs. 1, 2, 3 and 4). However, electron microscopic photos, taken during co-cultivation studies revealed that Pandoravirus inopinatum virions or developmental stages were never included within these packages (Figs. 2 and 4). Neither digested, partially digested or viable (undamaged) virions were found within these packages.
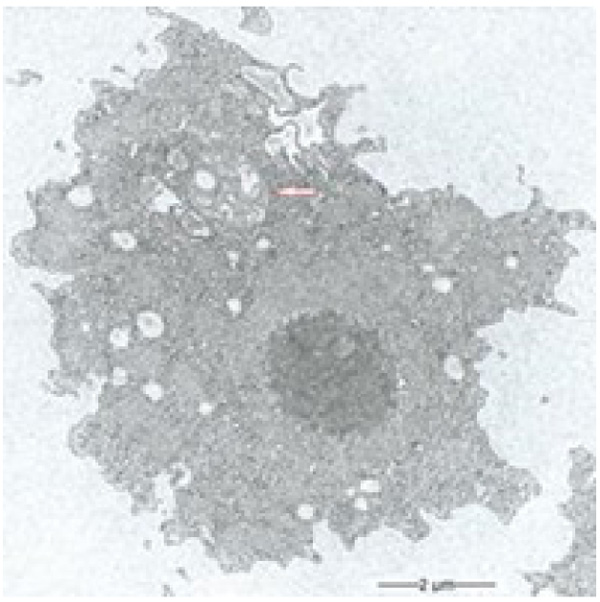
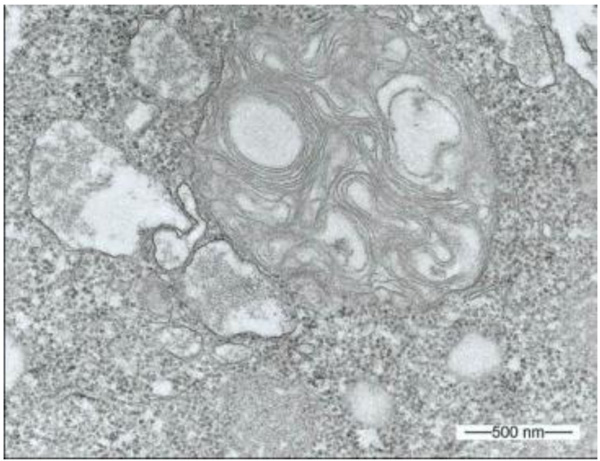
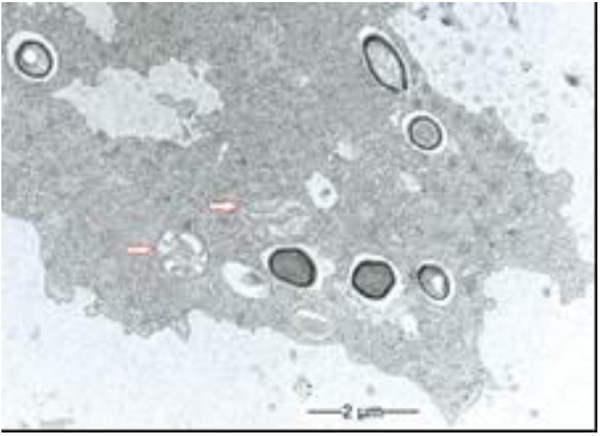
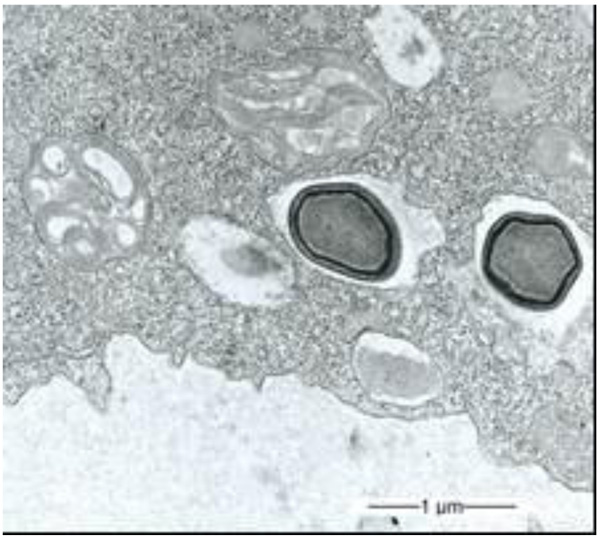
3.2. Results of the Host Range - Time Series Analysis
The results of the long term observation and time series analysis are summarized in Table 2 (modified host spectrum analysis). The uptake of the Pandoravirus virions and the subsequent replication turned out to be very similar in all axenic FLA cultures. The infection rate was higher compared to xenic cultures, which provided Enterobacter cloacae bacteria as food source. While only semi-quantitative results were achievable with such a study design, specific observations of the “co-cultivation” (more precisely: cultivation of FLA in presence of Pandoraviruses) of Pandoravirus inopinatum with potential host amoebae are included in Table 2 [14, 15].
| FLA (Acanthamoebae) | Strain | Specific Observations of the Amoeba - Pandoravirus Relationship; Proliferation Rate, in Comparison to the LaHel-KLaHel Findings |
|---|---|---|
| Acanthamoeba sp. | LaHel | Uptake of the virions immediatly after "co-cultivation"; after 2 days trophozoites were recognizable via light microscopy showing fulminante virus replication. One day later the trophozoites seem to be filled with virions only. |
| Acanthamoeba sp. | Maha | Longer adhesion times of the virions at the amoebal surface bevor uptake; proliferation in axenic and xenic cultures; 3 days after cocultivation viral particles inside the trophozoites. 7 days later trophozoites rounded up; |
| Acanthamoeba sp. | MaTa | Virusuptake in xenic culture immediatly after "co-cultivation" in axenic and xenic cultures; but limitate impact on the population; proliferation in axenic and xenic cultures; |
| Acanthamoeba sp. | TaHa | Fewer virus uptake compared to LaHel; immidiate virion uptake after "co-cultivation"; virion uptake in axenic cultures higher than in xenic cultures; encystation started from day 4 after cocultivation; trophozoites rounded up after 5 days; after 5 hours replication signs detectable; axenic similar |
| Acanthamoeba sp. | Petz | Similar to LaHel; immidiate virion uptake after "co-cultivation", higher encystation rate compared to LaHel; axenic similar; rounding up after three days |
| Acanthamoeba sp. | Ulse | Uptake starts 1,5 hours after "co-cultivation"; replication similar to Acanthamoeba sp. strain LaHel, the virions remained longer in the food vacuoles. I took three days until the virus particles could be detected within the cytoplasm of the amoebal trophozoites. A delayed developmental phase could be stated, compared to the development of Pandoravirus inopinatum in Acanthamoeba sp. strain LaHel. Apoptosios – like processes seemed to be involved, resulting in an increase in degraded or vacuolized trophozoites to prevent virus replication. |
| Acanthamoeba sp. | Ubo | Proliferation of Pandoravirus inopinatum within Acanthamoeba sp. Strain UBO only in the axenic medium; uptake similar to LaHel; uptake visible one day after cocultivation; virions visable; xenic: uptake could be no replication visable; |
| Acanthamoeba sp. | Flobö | Uptake of the virions visable after 1,5 hours after "co-cultivation"; the increased encystation rate seemed to be a result of the host - virus interaction; after 4-5 days incresed cell lysis; this strain was particularly suitable to document Pandoravirus virions within amoebal cysts. |
| Acanthamoeba sp. | KLB | Uptake similar; No replication signs in xenic medium; slow growing Acanthamoebae; axenisation not successful |
| Acanthamoeba sp. | Smith | Uptake immediatly; replication similar to KLaHel; three days after "Co-cultivation" many ruptured trophozoites; |
3.3. Specific Observations of the Host - Endocytobiont Relationship
By using the light microscopical time series analysis, the mode (time, manner) of uptake and transport of Pandoravirus inopinatum virions by the trophozoites of the putative host amoebae could be demonstrated clearly. The virions were taken up after food cup building via phagocytosis and transported initially in food vacuoles in the cytoplasm. These developmental features were achieved and archived for all the suitable host amoebae. As an example the uptake of a Pandoravirus inopinatum virion by a trophozoite of Acanthamoeba sp. strain Petz, is documented in Fig. (5).
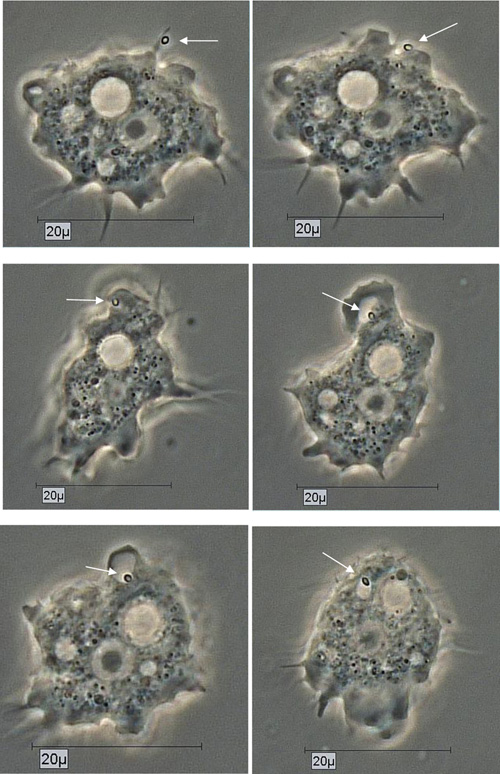
Due to their enormous size, the presence of Pandoravirus inopinatum virions within the host amoebae could be clearly shown by light microscopic investigations. The virions were detected in the trophozoites (Fig. 6) and the cysts of the host amoebae (Fig. 7)

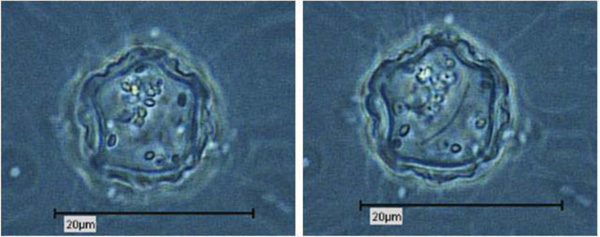
4. DISCUSSION
4.1. Packaging and Forming of Multilamellar Bodies
Endocytobionts ( e.g. bacteria) escape digestion by inducing host cell lysis or by exocytosis as free microorganisms, or packaged in the egested pellets [6, 19]. Occasionally bacterial endocytobionts are packaged into multilamellar bodies of lysosomal origin, which are formed by several concentric layers of lipid membranes and then excreted into the “environment” [20]. Acanthamoeba spp. and several species of ciliates are able to package viable (bacterial) pathogens, including Legionella pneumophila, Salmonella enteritica, Listeria monocytogenes, and Escherichia coli O157:H7 into multilamellar bodies [13, 21]. This FLA-driven process of packaging enables those bacteria to be resistant to digestion and subsequently to persist in the (unfavorable) environment, resulting in an enhancement of the survival of bacteria. The packaging by FLA (and other protozoa) is therefore a process, contributing to the enhanced survival of non-digested bacteria and dispersal of (pathogenic) bacteria in the environment. For example, Salmonella enteritica bacteria, when packaged in multilamellar bodies by the ciliate Tetrahymena, are more resistant to low concentrations of calcium hypochlorite compared to their planktonic state. Salmonella enteritica can even multiply inside these pellets [19, 22]. In addition to several other factors [7, 23], this process may be another source of pathogenic bacteria which could explain many infections, including some of the respiratory tract [13]. In the case of those respiratory pathogens, the packed bacteria can penetrate (as aerosols) deeper into the respiratory tract, cause respiratory infections, and remain undetected by the immune system, a fact that leads to an increase in their public health significance [13].
The packages are membrane-enclosed multilamellar bodies, composed of concentric membrane layers [24]. Multilamellar bodies are round to oval lysosomal cell organelles, enveloped by a double membrane, containing multiple thin electron dense lamellae. The presence of multilamellar bodies is associated with various lysosomal storage diseases (including gangliosidosis, Tay-Sachs syndrome, morbus Fabry and Niemann-Pick syndrome), associated with deficiencies in various lysosomal degradative enzymes and aberrant lysosomal accumulation of lipids [25].
Multilamellar bodies have been found in various cell types, where their principal functions are storage and secretion of lipids [26]. Studies have included not only the naked FLA but also the social amoeba (slime mould) Dictyostelium discoideum [5]. The formation of the multiple membrane lamella of the multilamellar bodies requires a vast amount of cellular lipids [24]. The lamellae are formed by repetitive inward budding of the membrane of the amoebal-lysosomal compartments [20]. The lipid composition of the multilamellar structures suggests that the pellets are not used only to expel undigested material: Lysosomal membrane proteins (including cysteine proteinase) were also detected in secreted expelled vesicles. In addition, undigested bacterial constituents or whole non-digested and digestable microorganisms are included in these multilamellar bodies.
Autophagy seems to be associated with the production of multilamellar bodies, although many elements of these processes still need to be clarified [13, 24, 27]. Autophagy is a normal degradative process, which exists in all eukaryotic cells and is stimulated in response to a variety of environmental stresses, which necessitate the use of autophagic mechanisms to enable cellular survival. It means that multilamellar bodies are formed via autophagic vacuole degradation. A specific role for autophagic sequestration in multilamellar bodies biogenesis was demonstrated [24].
The conditions which favor the production of packaged bacteria and their distribution in natural environments or man-made structures are not sufficiently known, whereas environmental conditions are believed to influence the packaging process [13] e.g. starving conditions are not the sole trigger for the production of multilamellar bodies. Nevertheless descriptive studies are available and achievable to state whether certain endocytobionts are involved in packaging processes.
Due to the unusual and enormous size of the Pandoravirus inopinatum virions (comparable to that of bacteria) we hypothesized that these virions could be expelled or otherwise involved in the packaging, as are bacteria. The electron microscopic photos were checked thoroughly for the presence of Pandoravirus inopinatum virions or developmental stages associated with the multilamellar bodies. Similar to the involvement of bacteria, and the packaging and the forming of multilamellar bodies, it seemed scarcely possible that endocytobionts of the size of >1µm could be secreted by the FLA, packed in multilamellar bodies. Multilamellar bodies were found frequently either within the Acanthamoeba trophozoites or expelled into the surrounding environment. While it is known that Acanthamoeba spp. are able to package viable (bacterial) pathogens into multilamellar bodies, there is no hint of Pandoravirus inopinatum stages being packed or involved at any stage in the packaging process at all [13, 21]. Therefore it can be concluded that the Pandoravirus-particles haven`t been part of this process and are therefore not affected (or protected) by these multilamellar structures.
4.2. Advanced Host Range Evaluation Method
Impressive light microscopic photos are available which were taken while observing the development of Pandoravirus inopinatum within Acanthamoebae, due to the fact that the Pandoravirus virions are about 1µm in size [10]. Light microscopy (completed by several stainings) can therefore be used for time series investigations and host-endocytobiont interaction studies in such cases. The uptake and the development of Pandoravirus virions was discussed and described by Scheid et al. [6, 10] and Abergel et al. [28]. The involvement of a virus factory in the development of Pandoraviruses was documented in 2015 [8].
The uptake of the virions could be demonstrated and documented for every potential host amoebal strain separately, using long term observations (Fig. 5). Virus replication could be detected in all co-cultivation studies, either in axenic co-cultivations or xenic co-cultivations or both. The 10 used Acanthamoeba strains showed differences in infectivity, susceptibility of Pandoravirus inopinatum, but they all turned out to be susceptible to Pandoravirus inopinatum. A correlation between the virus uptake and/or replication and the affiliation to T groups was not detected. Most of the used Acanthamoeba host strains belonged to the T 4 genotype; one belonged to T 11. Therefore the host specificity is not limited to the genus, the species, or even the genotype. The differences showed up only in the rate of virion uptake and replication, which demonstrated that the size of the trophozoites did not correlate with the replication rate: The replication rate of Pandoravirus inopinatum in Acanthamoeba strain Smith (a strain with smaller trophozoites) was higher than with Acanthamoeba sp. Strain HLA (the strain with the largest trophozoites in this study).
While only semi-quantitative results were achievable with the chosen study design, specific observations of the “co-cultivation” of Pandoravirus inopinatum with potential host amoebae were possible. With respect to the overall statement concerning the host range, a quantitative evaluation is not needed [14, 15]. Quantitative studies require qPCR-methods rather than microscopic evaluations. The use of a direct and dynamic microscopic examination as the imaging method, however, revealed several reaction types of Acanthamoebae against Pandoravirus inopinatum: Apoptotic processes, autophagy or increase of the encystment rate are defense reactions (Table 2). To determine the responsible genes or to quantify these defense mechanisms, biochemical studies are needed.
While there are limited results available using the “usual” host spectrum analysis methods (in most studies: does an uptake and/or replication of the endocytobiont occur at all?; results after several days concerning the amoebal culture), the process of infection or internalization can be studied specifically at one amoebal trophozoite for up to five hours with our method. This modified host spectrum analysis enables the investigation and documentation of intracellular processes. The study of interaction processes requires time lapse functions in contrast to static images by the usual light microscopic or electron microscopic approach, showing results at a certain moment, a snapshot of interaction.
CONCLUSION
The uptake of the Pandoravirus virions and the subsequent replication turned out to be very similar in all Acanthamoeba strains used as potential hosts. The infection rate was higher in axenic than in xenic cultures with Enterobacter cloacae bacteria as food source. The axenic culture is suitable for studies involving any quantifications concerning the endocytobiont proliferation, while studies to determine the viral replication rate by using real time qPCR methods are not performed easily [29, 30]. The use of further investigation measures such as distinct molecular biological methods will be more targeted and expedient. As the xenical culture is closer to real environmental conditions within soil or water in biofilms, both methods (Xenical and axenical culture) are justifiable and it has to be decided, which one is the preferred or expedient method to achieve specific valuable results leading to any realistic statements.
The detected differences in the uptake rate of axenical versus xenical cultures should be further investigated with respect to the gene expressions at certain stages. Biochemical and molecular biological methods (e.g. RNA sequencing) will certainly reveal details to determine finally whether an amoebal strain is a host with replication or a (transport) vehicle only (endozoochory) for these endocytobionts. Different genes seem to be activated in different environments (with different nutrition resources) resulting in different uptake rates and leading to different outcomes. Apoptosis-like processes or virustatic procedures were detected in xenic co-cultures. The rounding up of the trophozoites, the vacuolization, and the condensation of the chromatin might be signs of apoptosis as a result of the interaction with Pandoravirus inopinatum. The contribution of autophagy and apoptosis of the different Acanthamoeba strains should thus be investigated in more depth. Likewise, the investigation with respect to other countermeasures of the trophozoites, might highlight and reveal the differences in the interaction of diverse Acanthamoeba strains and Pandoravirus inopinatum. For example, so called “danger signals” (e.g. cytokines) are known in nature, especially from the immune systems of vertebrates [31].
Environmental conditions and nutritional factors ( e.g. peptone in the axenic medium available) seem to have an exceptional influence on the amoebae with respect to their interaction with Pandoravirus inopinatum virions. Differences in the amount of uptake, the uptake rate, the cyst-building process (and rate) and the replication proved that the in vitro conditions are critical for the determination and assessment of the susceptibility and the host spectrum, which in turn can be helpful to find the environmental main host. Therefore it is important to declare the specific in vitro conditions and avoid generalizations, as a small difference in any of these conditions may alter the results.
Initial studies to determine the host spectrum of Pandoravirus inopinatum were restricted to the fact of whether there was an uptake and replication or not. In general the time series results are more accurate and provide better kinetic details. Detailed statements as to the infection rate, replication rate and further interactions are available by the time series investigation. Although this method is more complex and time consuming, it reveals better and more relevant results compared to the usually performed host spectrum analysis. Therefore this modified version should replace the traditional (static) host spectrum analysis method to achieve deeper and more accurate insights into host - endocytobiont interactions.
A suitable (not human pathogenic) endocytobiont might be used as a targeted countermeasure against Acanthamoeba in the future (whether in soil, water, or to treat an Acanthamoeba keratitis?). The destruction of the trophozoites while replicating, the induction of specific biochemical processes, or the induction of apoptosis might be helpful tools in the future to face the risk of FLA as human pathogens. This approach still needs a thorough investigation before considering pandoraviruses as therapeutic microorganisms when considering them as applicable biological control agents against Acanthamoeba, e.g. in a keratitis patient or in limited natural areas, in soil and in aquatic habitats. Nevertheless, a selective and well-aimed application of parasitic endocytobionts ( e.g. Pandoraviruses seems to be expedient and promising) which turned out to be non - pathogenic and non - toxic for humans and animals. The confirmed suitable host range against Acanthamoebae enables Pandoraviruses to be used to reduce or control several pathogenic Acanthamoeba strains. The reduction of those Acanthamoebae in their role as hosts of pathogenic endocytobionts ( e.g. legionellae) could provide an additional, indirect benefit. These Pandoraviruses might be capable of keeping a certain environmental area free of Acanthamoebae, which is especially important when these defined areas, e.g. soil around hot (thermal) springs, are related to human activity.
To date there are no existing studies of the targeted application of amoeba destroying endocytobionts or giant viruses (giruses) in natural risk areas with confirmed pathogenic Acanthamoeba populations. The monitoring of interactions with Acanthamoebae results exclusively from in vitro studies. Possibly, the combination of two or more biological control agents ( e.g. two endocytobionts or non- human pathogenic giant viruses plus amoebophhagous fungi simultaneously) could enhance their activity to decimate the Acanthamoebae or at least reduce them below a “damage threshold”. The real effectiveness of Pandoraviruses with respect to the reduction of Acanthamoeba populations in natural habitats needs verification by means of field studies. But it is certainly worth considering, since such a biological control method could provide long - term and sustained effects.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGMENTS
The author would like to thank Dr. David Lam (MD, MPH, Shaman Medical Consulting) for review and English language editing of the article and Dr. B. Hauroeder and her team for the help to obtain the impressive electron microscopical records.


